I did, you never addressed it in our formal debate and your debate tactics haven't improved in all the time since.
Snide rhetorical jabs do not help your "case".
(Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 2005)
Seems like we did this before, you want everyone to believe that almost 8% of the human genome is the result of dangerous germline invasions.
It's what the researchers conclude from the full genome surveys.
With more than 100 members, CERV 1/PTERV1 is one of the most abundant families of endogenous retroviruses in the chimpanzee genome. CERV 1/PTERV1 elements range in size from 5 to 8.8 kb in length, are bordered by inverted terminal repeats (TG and CA) and are characterized by 4 bp TSDs...The age of each subfamily was estimated by calculating the average of the pairwise distances between all sequences in a given subfamily. The estimated ages of the two subfamilies are 5 MY and 7.8 MY, respectively, suggesting that at least one subfamily was present in the lineage prior to the time chimpanzees and humans diverged from a common ancestor (about 6 MYA). This conclusion, however, is inconsistent with the fact that no CERV 1/PTERV1 orthologues were detected in the sequenced human genome. (
Retroelements and the human genome: New perspectives on an old relation)
This indicates the Chimpanzee is being inundated by ERVs with over a million base pairs becoming permanently fixed right at the time of the split. ERV germline invasions are rare at best (
evidence?) and to have so many, many with open reading frames is a formula for extinction. Which means you ignore the differences and overstate things in common. What you are attempting to do is evade in the inverse logic.
Really don't see the point, let's move on.
If you don't see the point, don't move on. Ask for clarification of whatever you don't understand.
Well you almost characterized them and little else. It's odd that your saying they are all complete (
this is a blatant lie) when the ones in the human genome are so riddled in mutations that they are no longer functional.
I told you that ERVs are largely nonfunctional due to mutations. Try to keep up.
Starting to see quotes without source material, not a good sign. (
A link is provided.)
Does your blog link come with a bibliography?
All relevant sources are provided. Sometimes they are a couple of links away. The blog is only an educational piece. You need to study it carefully if you doubt its accuracy.
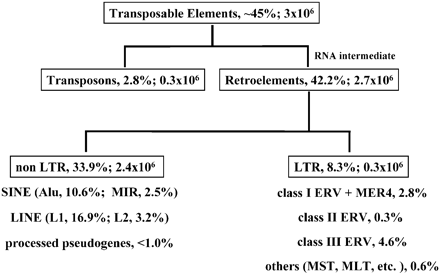
What they found in the Initial Sequence of the Human Genome. Their estimates apply only to the human genome, comparisons wouldn't come for several years:
112,000 ERV class 1 79.2 Mb 3.69% of the genome
8 ERV class II 8,9 Mb .31%
83,000 ERV class III 39.5 Mb 1.44%
Table 11. Number of copies and fraction of genome for classes of interspersed repeat.
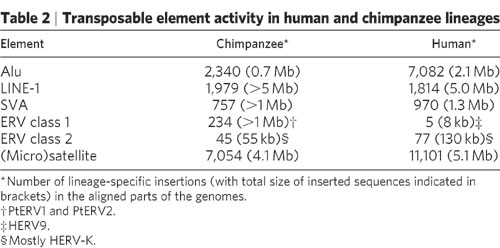
These would not be compared to the chimpanzee genome until 2005. They found 73 human-specific insertions of the HERV-K and 45 chimpanzee-specific insertions, leaving ~9 human-specific insertions before dying out. This comparison put's the amount of DNA from retroviruses at 5.44%. In the Chimpanzee genome it's almost 8%, 'With more than 100 members, CERV 1/PTERV1 is one of the most abundant families of endogenous retroviruses in the chimpanzee genome.' (Nature 2005).
Against this background, it was surprising to find 234 ERV class 1 > then a million base pairs in the Chimpanzee genome but absent in human DNA:
Notice the ERV class 1 and this discussion from the Chimpanzee Genome paper:
Against this background, it was surprising to find that the chimpanzee genome has two active retroviral elements (PtERV1 and PtERV2) that are unlike any older elements in either genome; these must have been introduced by infection of the chimpanzee germ line. The smaller family (PtERV2) has only a few dozen copies, which nonetheless represent multiple (~5–8) invasions, because the sequence differences among reconstructed subfamilies are too great (~8%) to have arisen by mutation since divergence from human.
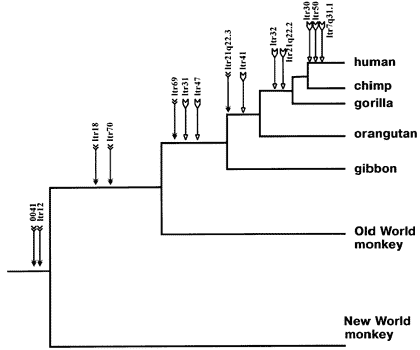
Figure 4.4.1. Human endogenous retrovirus K (HERV-K) insertions in identical chromosomal locations in various primates
There are at least seven different known instances of common retrogene insertions between chimps and humans, and this number is sure to grow as both these organism's genomes are sequenced (
see Prediction 4.5: Molecular evidence - Endogenous retroviruses)
The essence of the argument is the strongest argument against germline invasions in the first place. They are rare and they are pretty much random. Once they hook you with the assumption that they must be the result of a germline invasion they use the probability argument for common ancestry. The only way it would be possible for mutations and insertions to be in the same place is a common ancestor. It's just another homology argument buried in highly technical viral research but it really comes down to a false assumption and some anecdotal comparisons.
The odds of a single germline invasion from an ERV is itself, astronomical. (
Evidence?) The very statistical argument my opponent is trying to make is the reason the first assumption is patently false.
We've done this, long technical discussions of ERVs doesn't help your case, it buries it. When your actaully interested in doing the reading and coming to an informed conclusion I'll be waiting, but not holding my breath.
Have a nice day

Mark