The slightly deleterious model I was working from had nothing about less effective repair mechanisms in it. It was about lowered efficiency of natural selection, not about changes in the mutation rate.
A mutation rate of 2.3 x 10^-8 per generation for each base pair in the genome (for substitutions) predicts the following. Humans are separated from the human/chimpanzee common ancestor by roughly 350,000 generations, and there are 3 x 10^9 base pairs in the genome, so we should expect to find 2.3 x 10-8 x 3 x 10^9 x 3.5 x 10^5 = 24 million accumulated mutations in humans in that amount of time. Add the same number in chimpanzees, and that mutation rate predicts 48 million single-base differences. That's a little higher than the 35 million observed single-base subsitution differences between humans and chimps (not surprising, since that mutation rate is at the upper end of estimates), but certainly in the right ball park. And the error is in the wrong direction, as far as you're concerned: the known mutation rate is more than enough to explain the observed differences between humans and chimps, not hopelessly inadequate, as you claim.
That's not how I'm getting it:
Table 3. Estimates of mutation rate assuming different divergence times and different ancestral population sizes
4.5 mya, pop.= 10,000 mutation rate is 2.7 x 10^-8
4.5 mya, pop.= 100,000 mutation rate is 1.6 x 10^-8
5.0 mya, pop.= 10,000 mutation rate is 2.5 x 10^-8
5.0 mya, pop.= 10,0000 mutation rate is 1.5 x 10^-8
5.5 mya, pop.= 10,000 mutation rate is 2.3 x 10^-8
5.5 mya, pop.= 10,000 mutation rate is 1.4 x 10^-8
6.0 mya, pop.= 10,000 mutation rate is 2.1 x 10^-8
6.0 mya, pop.= 100,000 mutation rate is 1.3 x 10^-8
Calculations are based on a generation length of 20 years and average autosomal sequence divergence of 1.33%
Table 4. Estimates of mutation rate for different sites and different classes of mutation
Transition at CpG mutation rate 1.6 x 10^-7
Transversion at CpG mutation rate 4.4 x 10^-8
Transition at non-CpG mutation rate 4.4 x 10^-8
Transversion at non-CpG mutation rate 5.5 x 10^-9
All nucleotide subs mutation rate 2.3 x 10^-8
Length mutations mutation rate 2.3 x 10^-9
All mutations mutation rate 2.5 x 10^-8
Rates calculated on the basis of a divergence time of 5 mya, ancestral population size of 10,000, generation length of 20 yr, and rates of molecular evolution given in Table 1.
(Estimate of the Mutation Rate per Nucleotide in Humans Michael W. Nachmana and Susan L. Crowella Genetics, 297-304, September 2000)
Now I know that at the time this was written the differences in the DNA were thought to be less then 2%. When this percentage doubles it only makes sense that the mutation rate increases. Notice that the indel mutation rate is estimated as well as the overall mutation rate by nucleotide.
Do the same calculation for the indels. The estimated number of indel mutations should be 2.3 x 10^-9 x 3 x 10^9 x 3.5 x 10^5 = 2.4 million predicted indels in humans, and an equal number predicted in chimpanzees, for ~5 million predicted indel differences between them. The observed number of indel differences is (as you have pointed out several times) 5 million, exactly as predicted.
That's the number of indels, when the size of the mutation increases then the mutation rate is supposed to increase with it is it not?
The fact that common descent makes exactly the the right prediction about indels is not a good argument against common descent, Mark. (Personally, I don't make a lot from this prediction, since the uncertainty on the estimated indel mutation rate should be very large -- but I sure don't see any signs of a problem for evolution here.)
I never once thought, much less argued that TOE was somehow threatened by 6% of the DNA being different. It is very curious though, when you realize that the percentage of base pairs in a genome is 3%-4% higher then previously thought don't you recalculate the mutation rate? I only ask because no one seems to be doing that.
So let's try again, Mark. What is the problem with the overall number of indel differences between humans and chimpanzees?
It's the fact that you are using the same mutation rate you did when you thought that the DNA was 99% the same.
The mutation rate you quoted above does just fine here. When are you going to get around to spelling out your problem?
When you tell me how this changed the statistical average.
The difference between 35 million base pairs and 145 million base pairs is that the latter includes mutations that affect more than one base. To focus on indels, since that's supposed to be what we're talking about, there are ~77 million base pairs that differ between humans and chimpanzees in a total of 5 million individual insertions and deletions. So indel mutations must occur about one-seventh as often as single-base substitutions (5 million / 35 million), and the average indel is about 16 base pairs long (with most indels being very short but rare ones being very long indeed). Still looking for that problem, Mark.
I was looking through the only textbook I have available to me citing medical evidence on mutations. It says:
"Gene mutations are relatively rare but overall 1% of infants have a single mutant gene 10-8 to 10-6 per locus per gamete. "
(Genetics in Obstentrics and Gynecology, 3rd ed. Simpson and Elias)
What is the problem? For one thing mutations are rare to begin with, it is very rare for them to become a permanent part of the genome and extremely rare for them to be adaptive on an evolutionary scale.
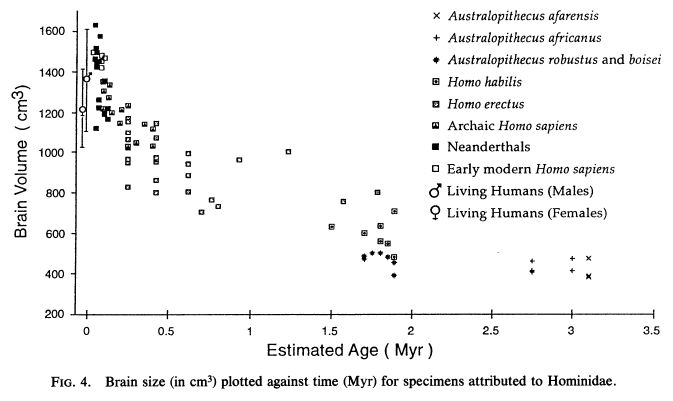
My problem is that the human brain is close to three times the size of a chimpanzee's. I also have a problem with random mutations being an explanation when the deleterious effects of mutations of fitness are the most likely effect from mutations.
I'm just curious, what would you say the difference between orthologous nucleotide sequences in their complement of genes, in the two genomes is?
Our results imply that humans and chimpanzees differ by at least 6% (1,418 of 22,000 genes) in their complement of genes, which stands in stark contrast to the oft-cited 1.5% difference between orthologous nucleotide sequences. (The Evolution of Mammalian Gene Families, Jeffery P. Demuth, Tijl De Bie, Jason E. Stajich, Nello Cristianini, and Matthew W. Hahn PLoS ONE.December 20, 2006)
The truth is coming out, I don't know or really care if this upsets the common ancestor model or not. The truth is I feel I'm wasting my time and it has nothing to do with genetics. I don't think you can ever really put a dent in someone else's a priori assumptions and common ancestry is no exception.
No, the mutation rate doesn't change. The ones you quoted above are still just fine. The substition rate is probably slightly high, but the indel rate is just about perfect (as I showed above) to give the 5 million observed indels.
So it really does not matter if it's a 1% difference or a 6% difference, the mutation rate is unchanged?
Note that to predict the number of base pairs of indel difference between humans and chimpanzees (rather than just the number of indels), you need to know the average size of an indel. The mutation rate estimate you quoted above doesn't do that, since it just describes the rate at which indel mutations occur, not how big they are.
All I can tell you for sure is that the bigger the indel the less likely it is to happen.
No, Mark, I don't know what your argument is. I'm sure that it's wrong, and that when I explain to you why it's wrong you'll again tell me I'm lying and that all evolutionary biologists are conspiring to conceal the obvious flaws in our own work, but we can't cycle through this idiotic process until you actually present the argument, can we? So could you present your argument already?
Steve, I'll level with you because I don't want to waste your time or mine. I'm tired buddy, I'm tired of looking for an answer to a question that has kept me into this for far too long. I simply want to know what it is that creates adaptations on a large scale. No one is interested and I'm tired of the drama.
Ok, you are telling me my arguments are idiotic and that is exactly what I am hearing from everyone. Don't worry about it because I'm just tired of trying. Three years of this and you guys still talk to me as if I were mentally retarded. I don't know what you are trying to accomplish but I finally found someone interested in a formal debate. I'm going to work on that and I think I'll just find something more productive to do with my time like beating my head against the wall.
That can't be the problem, since the quotation doesn't say that. 1.4% of the observed indels were over 80 bp in length (70,000 out of 5 million), which is not remotely the same as 73% being over 70,000 in length; those 1.4% contribute 73% of the sequence difference. In other words, there are lots of small indels (occurring at exactly the predicted rate, I'll note again), and a small number of large ones. And guess what? It turns out that small numbers of large indels also occur in modern humans. The mutation rate doesn't change, since the mutation rate counts the number of indels, not their size.
So try again: what's the problem with the indel rate?
No, we're not talking about those things. Those are the things you always try to change the subject to when I challenge you. What I'm doing is trying to get you to defend one very specific claim, which is that the indel differences between humans and chimpanzees are too large to be explained by evolution. First defend that claim.
Right now I would settle for someone admitting that the mutation rate changes when the differences go from 1% to 6%. Still in the interests of open discussion lets take another look at this:
"but that the largest few contain most of the sequence (with the approx70,000 indels larger than 80 bp comprising 73% of the affected base pairs)"
I really don't know what the problem was with what I said earlier and frankly I'm over it whatever it was. There are 70,000 indels that are larger then 80 base pairs. So that, according to the paper I wish I never read, it accounts for 73% of the affected base pairs. we are talking about 45 million base pairs in the human lineage.
That sounds like 1, 80 base pair indel, permanently fixed every 100 years. By the way, most of the changes had to occur around 2 million years ago. Homo habilis was little more then a big chimpanzee and Homo erectus was human in every way that is meaningful to me. This happened 2 mya pretty suddenly and if that is not a problem for you then your view of natural history is no longer a cause of concern for me.
You said that we should have found them, since we had the sequence of the human genome. And I want to know how having the sequence of the human genome would have let us find them. I still want to know.
I'm not sure what you mean but I have a headache and I have had a really weird day. I'm pretty disgusted with CF right now and while it has been the highlight of my time on here to discuss these things with you I think I've listened to as much of this as I can stand.
I was pretty much buying that we were very close to the chimpanzee in our DNA when I happened upon the Chimpanzee Chromosome 22 paper. It raised some questions so I pursued it. Then the Chimpanzee Genome paper came out and when I started this business of mutation rates and differences I was just curious how it changed the mutation rate.
You keep saying it doesn't, for me that indicates I am wasting my time with this.
I do owe you guys a lot though, you will never know just how frustrating it is to try to learn about genetics from Creationist essays. The only reason I stuck with this for so long is because I thought there was a chance that I might get other creationists interested in exploring the life sciences. It sounds kind of stupid to me now but that was the only thing that was really holding my interest.
I'll pop in from time to time and see if you guys have any comments on the formal debate but I don't see any reason to pursue this any further beyond that.
Straight up Steve, I deeply appreciate your efforts and patience. Maybe you guys are right and I'm just incredulous or something, I don't know. Either way I'm going to find something else to do with my time.
Grace and peace,
Mark