- Mar 16, 2004
- 22,030
- 7,265
- 62
- Gender
- Male
- Faith
- Calvinist
- Marital Status
- Single
- Politics
- US-Democrat
I spend most of my time in the Origins Theology forum but theistic evolutionists have little patience of reading scientific literature. I think the reason must be that you think very differently about science then you do religion. Which makes it hard to get them into the details of something as intricate as Genomics. I have also promised LM a discussion of ERVs that he considers a virtual smoking gun for Chimpanzee/Human common ancestry. At any rate, the purpose of the thread is to take up the subject of comparative genomics.
There is a standard line in the Darwinian theater of the mind that we are 98% the same as the Chimpanzee in our DNA. The scientific world has known for some time that this isn't true but you still keep seeing this in their popular press.
The fact is that we are not 98% the same in our DNA:
The paper cites these references:
It is puzzling that the announcement of the article would make this obviously bogus statement:
Loudmouth and I had been debating and discussing for some time when I finally talked him into a formal debate. The formal debate thread had been started and I was itching to put it to good use. Most evolutionists would not dare let a creationist get then on record, losing a debate with a creationist would be academic suicide. Casual discussion boards is another matter. At any rate, the discussion of ERVs can be found here:
mark kennedy v. Loudmouth
It's an homology argument, basically it says that because we are so similar we must have a common ancestor. The subject matter was new to me so I thought I was going to have to do a lot of reading but the refutation was shockingly easy to find:
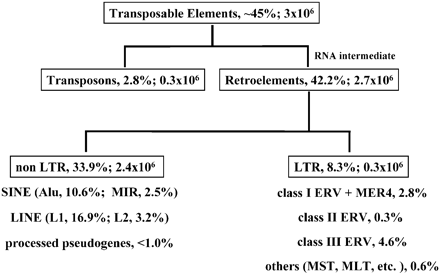
With more than 100 members, CERV 1/PTERV1 is one of the most abundant families of endogenous retroviruses in the chimpanzee genome. (Genome Biol. 2006). They can be found in African great apes but not in humans. What is more the ERV virus is nearly extinct in the human genome with only a couple that actually work. The only thing that ERVs are proof of is the lengths evolutionists will go to to conflate and confuse the evidence.
Ok, I know what your thinking, who care about difference in junk DNA right? What about protein coding genes? The Comparison of Human Chromosome 21 and Chimpanzee Chromosome 22 revealed that 83% of the 231 coding sequences, including functionally important genes, show differences at the amino acid sequence level. These included gross structural changes affecting gene products far more common than previously estimated (20.3% of the PTR22 proteins) (Nature, 27 May 2004). Genome wide the differences are now known to include 35 million single base substitutions, and five million indels (with the ~70,000 indels larger than 80 bp comprising 73% of the affected base pairs). This is in addition to 8 chromosomal rearrangements from 2 million base pairs to 4 million base pairs in length coming to more then 20 million base pairs.
One has to wonder what the mutation rate would have to be in order to get this kind of divergence. I expected a formula something like this one:
For reasons I can't quite grasp no evolutionist will touch the mutation rate with a ten foot pole. Now I managed to talk an MIT staff scientist into showing the rate for single base substitutions but they avoid the indels like the plague.
This is the one that sealed the deal for me. This one staggers the imagination and evolutionists literally have no clue how it could have happened:
Here's how it breaks down Barney style. For 400 million years this gene allows only 2 substitutions. In the lineage that became human it allows 18, how is this even conceivable, never mind possible and it is certainly not explained.
The thread is about homology arguments as they relate to comparative genomics. Like I said, I promised LM I would discuss the ERVs with him when I got some time and I think I can manage it now. Usually when I bring the subject up the average troll will avoid the thread like the plague, they are just not capable of doing this kind of technical reading. They lose the tag team effect which is their support system so they just peel off one by one.
Be advised I will not fall prey to fallacious circular arguments so if that's all you have don't blame me when you get your feeling hurt. I only mention that because I know what the trolling tactics are like on here and I don't want the novice trollers to get caught up in the mix. It is not my intention to discourage people with an interest in evolutionary biology to shy away from the scientific literature. On the contrary, I think these forums could go a long way to improve the ability of the layman to navigate these highly technical publications.
That's by no means an exhaustive sampling of the subject matter but you'll probably have to agree that it's a little more technical then the average thread on here. I also want to add that if you are a Christian who has embraced theistic evolution I'm perfectly willing to discuss the doctrinal and theological issues but I would rather do that in another thread, preferably in the Origins Theology forum.
Grace and peace,
Mark
There is a standard line in the Darwinian theater of the mind that we are 98% the same as the Chimpanzee in our DNA. The scientific world has known for some time that this isn't true but you still keep seeing this in their popular press.
The fact is that we are not 98% the same in our DNA:
On the basis of this analysis, we estimate that the human and chimpanzee genomes each contain 40–45 Mb of species-specific euchromatic sequence, and the indel differences between the genomes thus total ~90 Mb. This difference corresponds to ~3% of both genomes and dwarfs the 1.23% difference resulting from nucleotide substitutions (Nature, 1 September 2005)
The paper cites these references:
- Britten, R. J. Divergence between samples of chimpanzee and human DNA sequences is 5%, counting indels. Proc. Natl Acad. Sci. (2002)
- Frazer, K. A. et al. Genomic DNA insertions and deletions occur frequently between humans and nonhuman primates. Genome Res. (2003)
- Locke, D. P. et al. Large-scale variation among human and great ape genomes determined by array comparative genomic hybridization. Genome Res. (2003)
- Liu, G. et al. Analysis of primate genomic variation reveals a repeat-driven expansion of the human genome. Genome Res. 13, (2003)
- Yohn, C. T. et al. Lineage-specific expansions of retroviral insertions within the genomes of African great apes but not humans and orangutans. PLoS Biol. (2005)
It is puzzling that the announcement of the article would make this obviously bogus statement:
What makes us human? We share more than 98% of our DNA and almost all of our genes with our closest living relative, the chimpanzee. (Nature, Web Focus)
Loudmouth and I had been debating and discussing for some time when I finally talked him into a formal debate. The formal debate thread had been started and I was itching to put it to good use. Most evolutionists would not dare let a creationist get then on record, losing a debate with a creationist would be academic suicide. Casual discussion boards is another matter. At any rate, the discussion of ERVs can be found here:
mark kennedy v. Loudmouth
It's an homology argument, basically it says that because we are so similar we must have a common ancestor. The subject matter was new to me so I thought I was going to have to do a lot of reading but the refutation was shockingly easy to find:

With more than 100 members, CERV 1/PTERV1 is one of the most abundant families of endogenous retroviruses in the chimpanzee genome. (Genome Biol. 2006). They can be found in African great apes but not in humans. What is more the ERV virus is nearly extinct in the human genome with only a couple that actually work. The only thing that ERVs are proof of is the lengths evolutionists will go to to conflate and confuse the evidence.
Ok, I know what your thinking, who care about difference in junk DNA right? What about protein coding genes? The Comparison of Human Chromosome 21 and Chimpanzee Chromosome 22 revealed that 83% of the 231 coding sequences, including functionally important genes, show differences at the amino acid sequence level. These included gross structural changes affecting gene products far more common than previously estimated (20.3% of the PTR22 proteins) (Nature, 27 May 2004). Genome wide the differences are now known to include 35 million single base substitutions, and five million indels (with the ~70,000 indels larger than 80 bp comprising 73% of the affected base pairs). This is in addition to 8 chromosomal rearrangements from 2 million base pairs to 4 million base pairs in length coming to more then 20 million base pairs.
Our results imply that humans and chimpanzees differ by at least 6% (1,418 of 22,000 genes) in their complement of genes, which stands in stark contrast to the oft-cited 1.5% difference between orthologous nucleotide sequences. This genomic “revolving door” of gene gain and loss represents a large number of genetic differences separating humans from our closest relatives. (The Evolution of Mammalian Gene Families)
One has to wonder what the mutation rate would have to be in order to get this kind of divergence. I expected a formula something like this one:
Table 3. Estimates of mutation rate assuming different divergence times and different ancestral population sizes
4.5 mya, pop.= 10,000 mutation rate is 2.7 x 10^-8
4.5 mya, pop.= 100,000 mutation rate is 1.6 x 10^-8
5.0 mya, pop.= 10,000 mutation rate is 2.5 x 10^-8
5.0 mya, pop.= 10,0000 mutation rate is 1.5 x 10^-8
5.5 mya, pop.= 10,000 mutation rate is 2.3 x 10^-8
5.5 mya, pop.= 10,000 mutation rate is 1.4 x 10^-8
6.0 mya, pop.= 10,000 mutation rate is 2.1 x 10^-8
6.0 mya, pop.= 100,000 mutation rate is 1.3 x 10^-8
Table 4. Estimates of mutation rate for different sites and different classes of mutation
Transition at CpG mutation rate 1.6 x 10^-7
Transversion at CpG mutation rate 4.4 x 10^-8
Transition at non-CpG mutation rate 4.4 x 10^-8
Transversion at non-CpG mutation rate 5.5 x 10^-9
All nucleotide subs mutation rate 2.3 x 10^-8
Length mutations mutation rate 2.3 x 10^-9
All mutations mutation rate 2.5 x 10^-8
Rates calculated on the basis of a divergence time of 5 mya, ancestral population size of 10,000, generation length of 20 yr, and rates of molecular evolution given in Table 1.
Calculations are based on a generation length of 20 years and average autosomal sequence divergence of 1.33%
-----------------------------------------------------
Estimate of the Mutation Rate per Nucleotide in Humans (Michael W. Nachmana and Susan L. Crowella
Genetics, 297-304, September 2000)
4.5 mya, pop.= 10,000 mutation rate is 2.7 x 10^-8
4.5 mya, pop.= 100,000 mutation rate is 1.6 x 10^-8
5.0 mya, pop.= 10,000 mutation rate is 2.5 x 10^-8
5.0 mya, pop.= 10,0000 mutation rate is 1.5 x 10^-8
5.5 mya, pop.= 10,000 mutation rate is 2.3 x 10^-8
5.5 mya, pop.= 10,000 mutation rate is 1.4 x 10^-8
6.0 mya, pop.= 10,000 mutation rate is 2.1 x 10^-8
6.0 mya, pop.= 100,000 mutation rate is 1.3 x 10^-8
Table 4. Estimates of mutation rate for different sites and different classes of mutation
Transition at CpG mutation rate 1.6 x 10^-7
Transversion at CpG mutation rate 4.4 x 10^-8
Transition at non-CpG mutation rate 4.4 x 10^-8
Transversion at non-CpG mutation rate 5.5 x 10^-9
All nucleotide subs mutation rate 2.3 x 10^-8
Length mutations mutation rate 2.3 x 10^-9
All mutations mutation rate 2.5 x 10^-8
Rates calculated on the basis of a divergence time of 5 mya, ancestral population size of 10,000, generation length of 20 yr, and rates of molecular evolution given in Table 1.
Calculations are based on a generation length of 20 years and average autosomal sequence divergence of 1.33%
-----------------------------------------------------
Estimate of the Mutation Rate per Nucleotide in Humans (Michael W. Nachmana and Susan L. Crowella
Genetics, 297-304, September 2000)
For reasons I can't quite grasp no evolutionist will touch the mutation rate with a ten foot pole. Now I managed to talk an MIT staff scientist into showing the rate for single base substitutions but they avoid the indels like the plague.
This is the one that sealed the deal for me. This one staggers the imagination and evolutionists literally have no clue how it could have happened:
The 118-bp HAR1 region showed the most dramatically accelerated change, with an estimated 18 substitutions in the human lineage since the human–chimpanzee ancestor, compared with the expected 0.27 substitutions on the basis of the slow rate of change in this region in other amniotes. Only two bases (out of 118) are changed between chimpanzee and chicken, indicating that the region was present and functional in our ancestor at least 310 million years (myr) ago. No orthologue of HAR1 was detected in the frog (Xenopus tropicalis), any of the available fish genomes (zebrafish, Takifugu and Tetraodon), or in any invertebrate lineage, indicating that it originated no more than about 400 myr ago
Here's how it breaks down Barney style. For 400 million years this gene allows only 2 substitutions. In the lineage that became human it allows 18, how is this even conceivable, never mind possible and it is certainly not explained.
The thread is about homology arguments as they relate to comparative genomics. Like I said, I promised LM I would discuss the ERVs with him when I got some time and I think I can manage it now. Usually when I bring the subject up the average troll will avoid the thread like the plague, they are just not capable of doing this kind of technical reading. They lose the tag team effect which is their support system so they just peel off one by one.
Be advised I will not fall prey to fallacious circular arguments so if that's all you have don't blame me when you get your feeling hurt. I only mention that because I know what the trolling tactics are like on here and I don't want the novice trollers to get caught up in the mix. It is not my intention to discourage people with an interest in evolutionary biology to shy away from the scientific literature. On the contrary, I think these forums could go a long way to improve the ability of the layman to navigate these highly technical publications.
That's by no means an exhaustive sampling of the subject matter but you'll probably have to agree that it's a little more technical then the average thread on here. I also want to add that if you are a Christian who has embraced theistic evolution I'm perfectly willing to discuss the doctrinal and theological issues but I would rather do that in another thread, preferably in the Origins Theology forum.
Grace and peace,
Mark
Last edited: