For the third and final time, get the facts straight. These are the facts and stop wasting time with these pointless, outdated Talk Origins arguments that simply don't square with the facts:
A total of 95.8% of these sites were non-orthologous when compared between species. [/CENTER]
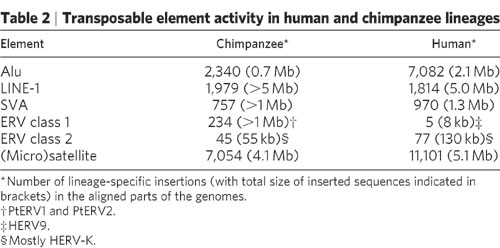
"We report here that the chimpanzee genome contains at least 42 separate families of endogenous retroviruses, nine of which were not previously identified. All but two (CERV 1/PTERV1 and CERV 2) of the 42 families of chimpanzee endogenous retroviruses were found to have orthologs in humans. Molecular analysis (PCR and Southern hybridization) of CERV 2 elements demonstrates that this family is present in chimpanzee, bonobo, gorilla and old-world monkeys but absent in human, orangutan and new-world monkeys. A survey of endogenous retroviral positional variation between chimpanzees and humans determined that approximately 7% of all chimpanzee-human INDEL variation is associated with endogenous retroviral sequences." (Identification, characterization and comparative genomics of chimpanzee endogenous retroviruses, Genome Biol. 2006 )
The most abundant family of ERVs in the Chimpanzee do not have ortholouges in the human genome:
With more than 100 members, CERV 1/PTERV1 is one of the most abundant families of endogenous retroviruses in the chimpanzee genome. CERV 1/PTERV1 elements range in size from 5 to 8.8 kb in length, are bordered by inverted terminal repeats (TG and CA) and are characterized by 4 bp TSDs...Phylogenetic analysis of the LTRs from full-length elements of CERV 1/PTERV1 members indicated that this family of LTRs can be grouped into at least two subfamilies (bootstrap value of 99; Figure 3). The age of each subfamily was estimated by calculating the average of the pairwise distances between all sequences in a given subfamily. The estimated ages of the two subfamilies are 5 MY and 7.8 MY, respectively, suggesting that at least one subfamily was present in the lineage prior to the time chimpanzees and humans diverged from a common ancestor (about 6 MYA). This conclusion, however, is inconsistent with the fact that
no CERV 1/PTERV1 orthologues were detected in the sequenced human genome.
(Identification, characterization and comparative genomics of chimpanzee endogenous retroviruses) Bolded mine


 That was the original intention of my OP! And we are four pages in and still haven't solved it.
That was the original intention of my OP! And we are four pages in and still haven't solved it.