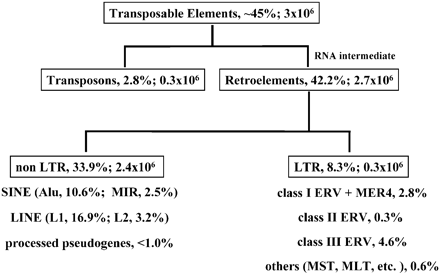
I've been corrected endlessly and you guys never get your facts straight. This was the first entire sequence of the human genome, it simply listed human ERVs, there would be nothing comprehensive regarding a comparison of them until 2005. By 2006 42 families of ERVs were identified, 40 of which had orthologues in the human genome and never are the ERVs described as identical, 2 had no orthologues in the human genome.
With more than 100 members, CERV 1/PTERV1 is one of the most abundant families of endogenous retroviruses in the chimpanzee genome…Consistent with our findings, the results of a previously published Southern hybridization survey indicated that sequences orthologous to CERV 1/PTERV1 elements are present in the African great apes and old world monkeys but not in Asian apes or humans . These results suggest that some members of the CERV 1/PTERV1 subfamily entered the chimpanzee…
…It has been estimated that 3.5% of the sequence differences between chimpanzees and humans is due to INDELs and that this INDEL variation may be of particular evolutionary significance. We have determined that approximately 7% of all chimpanzee-human INDEL variation is attributable to the presence or absence of endogenous retroviral sequences. (Identification, characterization and comparative genomics of chimpanzee endogenous retroviruses. Genome Biol. 2006)
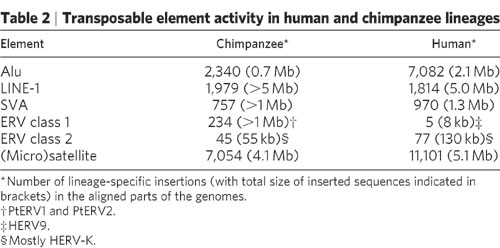
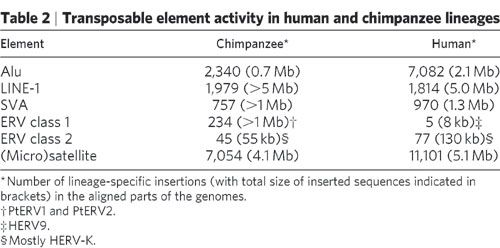
The CERV1/PTERV1 is one of the most abundant ERVs in the human genome and does not exist in the human genome. 3.5% of the differences are due to INDELS and 7% of the entire divergence due to INDELS. You call this an argument for common ancestry? ERV class 1 elements simply do not exist in the human genome and compare them to the class II and III elements:
In case you missed it that 234 in the Chimpanzee genome and 5 in the human genome. The class I elements are the PtERV1 and PtERV2 elements making this far and away the worst homology argument I've ever seen.