So much I could respond to here if I had the time, but in this post I will limit myself to this topic since you seem to be hot on it. You keep mentioning the huge change in brain size of hominid fossils after Lucy. How can we explain the large increase in brains with Homo habilis? Simple, Homo habilis needed to be smart to survive. OK, but didn't every animal need to be smart? Well, yes, but brains are expensive. They require enormous amounts of energy, and they are limited by the size of the head that can fit through the birth canal. So for most animals, they live with the size of the head they have, and that is that. But for Homo it was different. Their ancestors had been standing upright for years, and that freed their hands up to use tools. One needs to be smart to use tools in novel ways. And they had begun to lose their hair, allowing them to participate in prolonged hunting excursions without being exhausted in the hot African sun like other animals. With enough persistence, they could eventually chase down weak animals that were exhausted in the hot sun. But most importantly, they were developing complex societies that required communication. All this required brains. Homo needed big brains so they could truly understand each other. So there was strong evolutionary pressure to expand the brain.
One of the most striking features regarding hominids is that the cranial capacity is consistent with apes, just slightly bigger in some cases. We are not talking about limb proportions but rather the highly conserved genes related to neural functions. Most of the brain related genes would require a massive overhaul including HAR1f,SRGAP2 and 60 devo (brand new) brain related genes.
- HAR1F: Vital regulatory gene involved in brain development, 300 million years it has only 2 substitutions, then 2 million years ago it allows 18, no explanation how.
- SRGAP2: One single amino-acid change between human and mouse and no changes among nonhuman primates. accumulated as many as seven amino-acid replacements compared to one synonymous change. 6 known alleles, all resulting in sever neural disorder.
- 60 de novo (brand new) brain related genes with no known molecular mechanism to produce them.
The Taung Child, that replaced the Piltdown hoax, is a chimpanzee, so is Lucy. Leakey mentions the Piltdown skull in his book 'Adam's Ancestors':
'If the lower jaw really belongs to the same individual as the skull, then the Piltdown man is unique in all humanity. . . It is tempting to argue that the skull, on the one hand, and the jaw, on the other, do not belong to the same creature. Indeed a number of anatomists maintain that the skull and jaw cannot belong to the same individual and they see in the jaw and canine tooth evidence of a contemporary anthropoid ape.'
He referred to the whole affair as an enigma: In
By the Evidence he says 'I admit . . . that I was foolish enough never to dream, even for a moment, that the true explanation lay in a deliberate forgery.' (
Leakey and Piltdown)
The problem was that there was nothing to replace it as a transitional from ape to man. Concurrent with the prominence of the Piltdown fossil Raymond Dart had reported on the skull of an ape that had filled with lime creating an endocast or a model of what the brain would have looked like. Everyone considered it a chimpanzee child since it’s cranial capacity was just over 400cc but with the demise of Piltdown, a new icon was needed in the Darwinian theater of the mind. Raymond Dart suggests to Louis Leakey that a small brained human ancestor might have been responsible for some of the supposed tools the Leaky family was finding in Africa. The myth of the stone age ape man was born.
Ah, but there was that barrier to the head size that could come out of the birth canal. Just like computers were long limited by the infamous DOS 64K memory limit, Homo had the birth canal limit to brain size. At first hominids adapted by giving up much of the sense of smell, and having the brain concentrate on cognitive tasks instead. Neat trick, but it can only do so much. But Homo Habilis developed something new. The head and the brain of the infants began to expand after birth. This had its drawbacks. While the brain is developing, human babies are nearly helpless. It takes them a year to stand upright, and many years to be self sufficient. Other animal babies can stand within minutes of birth, and are soon ready to fend for themselves. But for Homo Habilis, the need for a larger brain was so strong, that it was worth going through a sustained period of helplessness as a child if they would end up smarter.
What they might need is completely beside the point, it's how this happens that matters. Mutations is brain related genes result in disease and disorder. Researchers have discovered at least two dramatic giant leaps that would have had to occur in order of the human brain to have emerged from ape like ancestors SRGAP2, HAR1F. In addition genes involved with the development of language (FOXP2), changes in the musculature of the jaw (MYH16) , and limb and digit specializations (HACNS1).
The ancestral SRGAP2 protein sequence is highly constrained based on our analysis of 10 mammalian lineages. We find only a single amino-acid change between human and mouse and no changes among nonhuman primates within the first nine exons of the SRGAP2 orthologs. This is in stark contrast to the duplicate copies, which diverged from ancestral SRGAP2A less than 4 mya, but have accumulated as many as seven amino-acid replacements compared to one synonymous change. (Human-specific evolution of novel SRGAP2 genes by incomplete segmental duplication Cell May 2012)
What is the problem with 7 amino acid replacements in a highly conserved brain related gene? The only observed effects of changes in this gene in humans is disease and disorder:
- 15,767 individuals reported by Cooper et al. (2011)] for potential copy-number variation. We identified six large (>1 Mbp) copy-number variants (CNVs), including three deletions of the ancestral 1q32.1 region…
- A ten year old child with a history of seizures, attention deficit disorder, and learning disabilities. An MRI of this patient also indicates several brain malformations, including hypoplasia of the posterior body of the corpus callosum…
- Translocation breaking within intron 6 of SRGAP2A was reported in a five-year-old girl diagnosed with West syndrome and exhibiting epileptic seizures, intellectual disability, cortical atrophy, and a thin corpus callosum. (Human-specific evolution of novel SRGAP2 genes by incomplete segmental duplication Cell May 2012)
You mention a number of genes that had to develop over a half million years for this increase in brain size. OK, not a big problem, mutations can certainly produce 60 significant changes in a half a million years. And where there is strong pressure for a particular feature, those mutations that have that feature will stay in the gene pool. So for habilis, having had an enormous need for larger brains, those genes were selected for. It is not unusual for a particular body part to double in size in a half million years if there is strong evolutionary pressure.
No, I'm not talking about 60 'significant changes', I'm talking about 60 de novo (brand new) brain related genes in additions to a major overhaul of brain related genes across the board.
“Is it a few mutations in a few genes, a lot of mutations in a few genes, or a lot of mutations in a lot of genes? The answer appears to be a lot of mutations in a lot of genes. We've done a rough calculation that the evolution of the human brain probably involves hundreds if not thousands of mutations in perhaps hundreds or thousands of genes—and even that is a conservative estimate.” (
Lahn, Human Evolution Was a 'Special Event'. Howard Huges Medical Institute)
60 de novo (brand new) brain related genes with no known molecular mechanism to produce them. Selection can explain the survival of the fittest but the arrival of the fittest requires a cause:
The de novo origin of a new protein-coding gene from non-coding DNA is considered to be a very rare occurrence in genomes. Here we identify 60 new protein-coding genes that originated de novo on the human lineage since divergence from the chimpanzee. The functionality of these genes is supported by both transcriptional and proteomic evidence. RNA– seq data indicate that these genes have their highest expression levels in the cerebral cortex and testes, which might suggest that these genes contribute to phenotypic traits that are unique to humans, such as improved cognitive ability. Our results are inconsistent with the traditional view that the de novo origin of new genes is very rare, thus there should be greater appreciation of the importance of the de novo origination of genes…(De Novo Origin of Human Protein-Coding Genes PLoS 2011)
Whatever you think happened one thing is for sure, random mutations are the worst explanation possible. They cannot produce de novo genes and invariably disrupt functional genes. You can forget about gradual accumulation of, 'slow and gradual accumulation of numerous, slight, yet profitable, variations' (
Darwin). That would require virtually no cost and extreme benefit with the molecular cause fabricated from vain imagination and suspended by pure faith.
This change was not overnight. Someone posted a chart earlier that showed the increase in brain size with time. But just like Moore's law took over when we exceeded the DOS 64K limit, nature took over when we began to exceed the limit imposed by the birth canal.
The human genome doesn't have an army of Microsoft research and development programmers producing regular upgrades. What you see is DNA repair mechanisms that maintain the genome, protecting against changes, especially in highly conserved genes since changes (mutations) will have dramatic and devastating consequences:
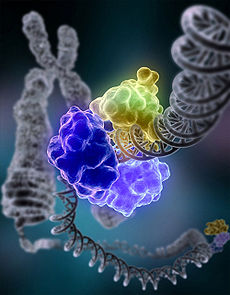
DNA ligase, shown above repairing chromosomal damage, is an enzyme that joins broken nucleotides together by catalyzing the formation of an internucleotide
ester bond between the phosphate backbone and the deoxyribose nucleotides. (
DNA Repair)
Back to transitionals. There is a broad range of transitional fossils leading up to humans. The brain size in adults get significantly bigger as one advances through the fossil record. See
Fossil Hominids: the evidence for human evolution .
Talk Origins has blown it's credibility with me sky high with this obviously erroneous statement:
The difference between chimpanzees and humans due to single-nucleotide substitutions averages 1.23 percent, of which 1.06 percent or less is due to fixed divergence, and the rest being a result of polymorphism within chimp populations and within human populations. Insertion and deletion (indel) events account for another approximately 3 percent difference between chimp and human sequences, but each indel typically involves multiple nucleotides. The number of genetic changes from indels is a fraction of the number of single-nucleotide substitutions (roughly 5 million compared with roughly 35 million). So describing humans and chimpanzees as 98 to 99 percent identical is entirely appropriate (Chimpanzee Sequencing 2005). (
Claim CB 144)
That defies the most basic math, the 5 million indels represents 90 million base pairs:
On the basis of this analysis, we estimate that the human and chimpanzee genomes each contain 40–45 Mb of species-specific euchromatic sequence, and the indel differences between the genomes thus total ~90 Mb. This difference corresponds to ~3% of both genomes and dwarfs the 1.23% difference resulting from nucleotide substitutions. (
Initial Sequence of the Chimpanzee Genome, Nature 2005)
This had been known for years before the entire sequence was sequenced and the known divergence has remained at least 5% ever since:
Five chimpanzee bacterial artificial chromosome (BAC) sequences (described in GenBank) have been compared with the best matching regions of the human genome sequence to assay the amount and kind of DNA divergence. The conclusion is the old saw that we share 98.5% of our DNA sequence with chimpanzee is probably in error. For this sample, a better estimate would be that 95% of the base pairs are exactly shared between chimpanzee and human DNA. In this sample of 779 kb, the divergence due to base substitution is 1.4%, and there is an additional 3.4% difference due to the presence of indels. (Divergence between samples of chimpanzee and human DNA sequences is 5%, counting indels. PNAS 2002)
They have no explanation for indels, a fair number over a million base pairs long.
FIGURE 6. Length distribution of large indel events (> 15 kb), as determined using paired-end sequences from chimpanzee mapped against the human genome.
That in addition to HAR1F, SRGAP2 (6 known alleles, all resulting in sever neural disorder.), and 60 de novo (brand new) brain related genes with no known molecular mechanism to produce them. Microsoft doesn't get software upgrades because the operating system needs them, it gets them because there is an army of software programmers producing them. The human genome has no such research and development teams, normative adaptive evolution happens primarily by preserving the fidelity of DNA sequences. Changes in protein coding genes and regulatory genes result in disease and disorder, not adaptation on an evolutionary scale:
Among the mutations that affect a typical gene, different kinds produce different impacts. A very few are at least momentarily adaptive on an evolutionary scale. Many are deleterious...If adaptive mutations are rare, as seems to be the case, then rates of DNA sequence evolution are driven mainly by mutation and random drift, (Rates of Spontaneous Mutations, (Genetics 1998)
Indeed, mutations are the only explanation except it's the worst possible vehicle due to the devastating disease and disorder that would undoubtedly result.