Back in 2009, I posted an essay on my website that I had just finished on the three ways endogenous retroviruses provide evidence for common ancestry. Since then, I have continually updated itespecially the section on common creationist responses. So when a creationist blogger, named Jonathan M, recently announced on the Evolution News & Views website that he intended on responding to my essay, I was intrigued.
But my intrigue turned to disappointment, once his two-part response was published (p1, p2):

And to top it all off, even though Jonathan M. didn't even attempt to explain the hierarchical patterns, or why they corroborate one another (something I lamented the universality of in the essay itself), he nonetheless boldly concluded with; Unfortunately for Darwinists, however, the evidence for common ancestry is paper thin on the ground.
All this, and much more, is discussed in the thorough rebuttal I posted. And judging by the continuous revisions Jonathan M. has been making to his shrinking response, it is clear that he has taken notice. Thus far, he has removed his entire paragraph and his quote on syncytin, removed any mention of gene expression and its associated quote, and corrected the misattribution he copied from Pitman. A screen shot of Google Caches June 3rd archival record tells the tale.
The last sentence of my rebuttal conclusion should sum up my sentiments:
But my intrigue turned to disappointment, once his two-part response was published (p1, p2):
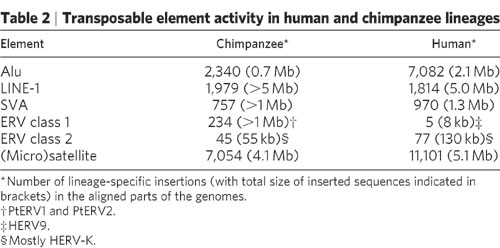
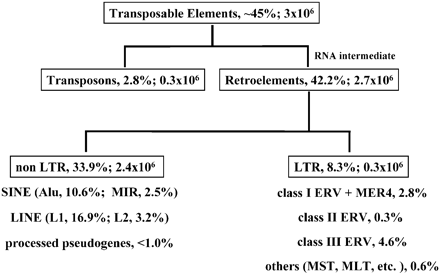
- It contained errors and misrepresentations, such as claiming that only a handful of the tens of thousands of ERVs shared between chimpanzees and humans are actually shared, presenting differences in expression between genes with and without intronic insertions as differences between genes with orthologous insertions, and classifying PtERV1 insertions as sources of deviation from the nested hierarchy of orthologues ERVs, rather than sources of deviation from the LTR-LTR discontinuity ratio patterns. It also contained what may be a functionality non sequitur, but what appeared to be the false claim that hominoid syncytin-1/2 and murid syncytin-A/B are in orthologous loci, respectively.
- Jonathan M. routinely presented arguments verbatim that were addressed in the common creationist responses section of the essay he claimed to be responding to. For instance, he presented the HERV-K-GC1 argument, where he concluded that the inserts in the chimpanzee and gorilla lineages must be independent events, without even addressing my extensive explanation of homologous recombination of duplicate genomic segments and allelic segregationboth of which were the focus of the research publication he was quoting from. He also repeatedly made the PtERV1 argument (mostly via uninterpreted quotes), which I had also explained in detail.
- Much of Jonathan Ms response was unoriginal, and the quotes were from abstracts of research publications he likely didnt read. Of the 9 quotes he provided, two were copy/pasted from a 2006 article by Dan Reynolds, and three were copy/pasted from a 2001 article by Sean Pitman (he also copied Pitmans misattribution of the quote to the wrong publication). Even the surrounding text he wrote sometimes read like a paraphrase. Yet despite all this, he never cited either of these articles as sources.

And to top it all off, even though Jonathan M. didn't even attempt to explain the hierarchical patterns, or why they corroborate one another (something I lamented the universality of in the essay itself), he nonetheless boldly concluded with; Unfortunately for Darwinists, however, the evidence for common ancestry is paper thin on the ground.
All this, and much more, is discussed in the thorough rebuttal I posted. And judging by the continuous revisions Jonathan M. has been making to his shrinking response, it is clear that he has taken notice. Thus far, he has removed his entire paragraph and his quote on syncytin, removed any mention of gene expression and its associated quote, and corrected the misattribution he copied from Pitman. A screen shot of Google Caches June 3rd archival record tells the tale.
The last sentence of my rebuttal conclusion should sum up my sentiments:
I can only hope that if anyone else responds to my formulation of the ERV argument for common ancestry, they do not provide non-responses, and do provide a "comprehensive working creationist model that is consistent with uncommon ancestry and incorporates the whole of ERV data."